install.packages("tidyverse")Pre-lecture materials
🌴
Read ahead
Before class, you can prepare by reading the following materials:
Acknowledgements
Material for this lecture was borrowed and adopted from
Learning objectives
At the end of this lesson you will:
- Understand the tools available to get data into the proper structure and shape for downstream analyses
- Learn about the dplyr R package to manage data frames
- Recognize the key verbs (functions) to manage data frames in dplyr
- Use the “pipe” operator to combine verbs together
Overview
It is still important to understand base R manipulations, particularly for things such as cleaning raw data, troubleshooting, and writing custom functions. But the tidyverse provides many useful tools for data manipuation and analysis of cleaned data. In this session we will learn about dplyr and friends.
Tidy data
The tidyverse has many slogans. A particularly good one for all data analysis is the notion of tidy data.
As defined by Hadley Wickham in his 2014 paper published in the Journal of Statistical Software, a tidy dataset has the following properties:
Each variable forms a column.
Each observation forms a row.
Each type of observational unit forms a table.

[Source: Artwork by Allison Horst]
What shapes does the data need to be in?
Beyond the data being tidy, however, we also need to think about what shape it needs to be in. Weʻll review concepts and tools in the next two lessons.
Now that we have had some experience plotting our data, we can see the value of having rectangular dataframes. We can also see that for particular graphics and analyses, we need to have the data arranged in particular ways.
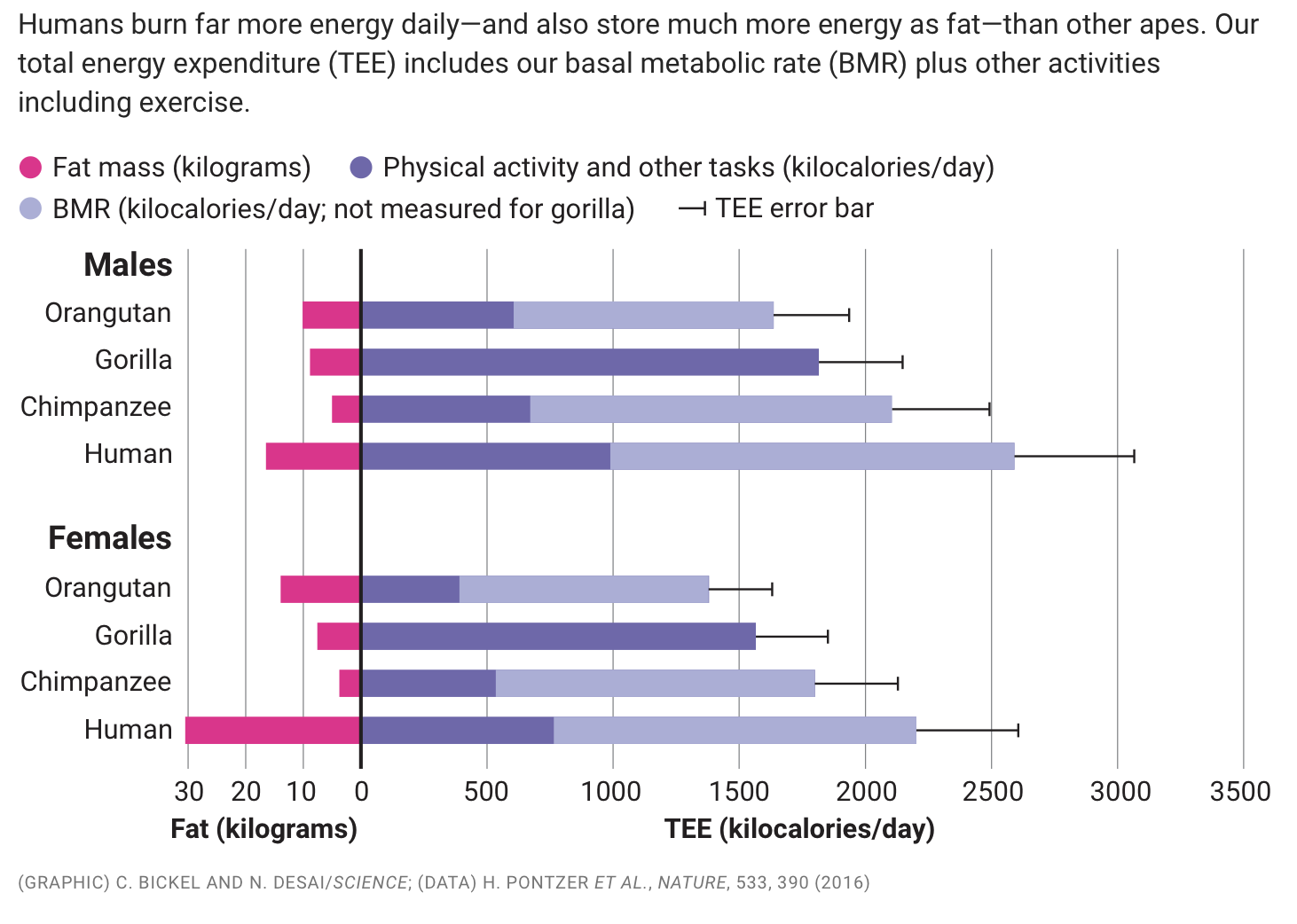
For example, take a look at this elegant graphic below. This single graphic is packed with information on fat, BMR, TEE, and activity levels, all for mulitple species. Is it more effective that individual bar plots? This arrangement is so helpful because you can imagine questions that can be answered with it by comparing the different aspects of the data.
- First imagine what it would look like variable by variable.
- How might you intially plot the data?
- What organization would you need to make a single figure such as this?
We often do not know exactly what we need at the start of a data analysis. We have to play around with different data structures, rearrange the data, look for interesting plots to try, rerrange to fit the input requirements of new functions weʻve discovered, and so on.
Tibbles
The tidyverse uses as its central data structure, the tibble or tbl_df. Tibbles are a variation on data frames, claimed to be lazy and surly:
- They don’t change variable names or types when you construct the tibble.
- Don’t convert strings to factors (the default behavior in
data.frame()). - Complain more when a variable doesnʻt exist.
- No
row.names()in a tibble. Instead, you must create a new variable. - Display a different summary style for its
print()method. - Allows non-standard R names for variables
- Allows columns to be lists.
However, most tidyverse functions also work on data frames. Itʻs up to you.
tibble() constructor
Just as with data frames, there is a tibble() constructor function, which functions in many ways with similar syntax as the data.frame() constructor.
If you havenʻt already done so, install the tidyverse:
Loading required package: tibbletibble( iris[1:4,] ) # the first few rows of iris# A tibble: 4 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa x <- 1:3
tibble( x, x * 2 ) # name assigned at construction# A tibble: 3 × 2
x `x * 2`
<int> <dbl>
1 1 2
2 2 4
3 3 6silly <- tibble( # an example of a non-standard names
`one - 3` = 1:3, # name = value syntax
`12` = "numeric",
`:)` = "smile",
)
silly# A tibble: 3 × 3
`one - 3` `12` `:)`
<int> <chr> <chr>
1 1 numeric smile
2 2 numeric smile
3 3 numeric smile
as_tibble() coersion
as_tibble() converts an existing object, such as a data frame or matrix, into a tibble.
as_tibble( iris[1:4,] ) # coercing a dataframe to tibble# A tibble: 4 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa As output
Most often we will get tibbles returned from tidyverse functions such as read_csv() from the readr package.
The dplyr package
The dplyr package, which is part of the tidyverse was written to supply a grammar for data manipulation, with verbs for the most common data manipulation tasks.

[Source: Artwork by Allison Horst]
dplyr functions
select(): return a subset of the data frame, using a flexible notationfilter(): extract a subset of rows from a data frame using logical conditionsarrange(): reorder rows of a data framerelocate(): rearrange the columns of a data framerename(): rename columns in a data framemutate(): add new columns or transform existing variablessummarize(): generate summary statistics of the variables in the data frame, by strata if data are hierarchical%>%: the “pipe” operator (from magrittr) connects multiple verbs together into a data wrangling pipeline (kind of like making a compound sentence)
Note: Everything dplyr does could already be done with base R. What is different is a new syntax, which allows for more clarity of the data manipulations and the order, and perhaps makes the code more readable.
Instead of the nested syntax, or typing the dataframe name over and over, we can pipe one operation into the next.
Another useful contribution is that dplyr functions are very fast, as many key operations are coded in C++. This will be important for very large datasets or repeated manipulations (say in a simulation study).
starwars dataset
We will use the starwars dataset included with dplyr. You should check out the help page for this dataset ?starwars.
Letʻs start by using the skim() function to check out the dataset:
Loading required package: dplyr
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionclass(starwars)[1] "tbl_df" "tbl" "data.frame"skimr::skim(starwars)| Name | starwars |
| Number of rows | 87 |
| Number of columns | 14 |
| _______________________ | |
| Column type frequency: | |
| character | 8 |
| list | 3 |
| numeric | 3 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| name | 0 | 1.00 | 3 | 21 | 0 | 87 | 0 |
| hair_color | 5 | 0.94 | 4 | 13 | 0 | 12 | 0 |
| skin_color | 0 | 1.00 | 3 | 19 | 0 | 31 | 0 |
| eye_color | 0 | 1.00 | 3 | 13 | 0 | 15 | 0 |
| sex | 4 | 0.95 | 4 | 14 | 0 | 4 | 0 |
| gender | 4 | 0.95 | 8 | 9 | 0 | 2 | 0 |
| homeworld | 10 | 0.89 | 4 | 14 | 0 | 48 | 0 |
| species | 4 | 0.95 | 3 | 14 | 0 | 37 | 0 |
Variable type: list
| skim_variable | n_missing | complete_rate | n_unique | min_length | max_length |
|---|---|---|---|---|---|
| films | 0 | 1 | 24 | 1 | 7 |
| vehicles | 0 | 1 | 11 | 0 | 2 |
| starships | 0 | 1 | 17 | 0 | 5 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| height | 6 | 0.93 | 174.36 | 34.77 | 66 | 167.0 | 180 | 191.0 | 264 | ▁▁▇▅▁ |
| mass | 28 | 0.68 | 97.31 | 169.46 | 15 | 55.6 | 79 | 84.5 | 1358 | ▇▁▁▁▁ |
| birth_year | 44 | 0.49 | 87.57 | 154.69 | 8 | 35.0 | 52 | 72.0 | 896 | ▇▁▁▁▁ |
Selecting columns with select()
Suppose we wanted to take the first 3 columns only. There are a few ways to do this.
We could for example use numerical indices:
names(starwars)[1:3][1] "name" "height" "mass" But we can also use the names directly:
# A tibble: 6 × 5
name sex gender homeworld species
<chr> <chr> <chr> <chr> <chr>
1 Luke Skywalker male masculine Tatooine Human
2 C-3PO none masculine Tatooine Droid
3 R2-D2 none masculine Naboo Droid
4 Darth Vader male masculine Tatooine Human
5 Leia Organa female feminine Alderaan Human
6 Owen Lars male masculine Tatooine Human The : normally cannot be used with names or strings, but inside the select() function you can use it to specify a range of variable names.
By exclusion
Variables can be omited using the negative sign withing select():
select( starwars, -(sex:species))The select() function also has several helper functions that allow matching on patterns. So, for example, if you wanted to keep every variable that ends with “color”:
tibble [87 × 3] (S3: tbl_df/tbl/data.frame)
$ hair_color: chr [1:87] "blond" NA NA "none" ...
$ skin_color: chr [1:87] "fair" "gold" "white, blue" "white" ...
$ eye_color : chr [1:87] "blue" "yellow" "red" "yellow" ...Or all variables that start with n or m:
subset <- select(starwars, starts_with("n") | starts_with("m"))
str(subset)tibble [87 × 2] (S3: tbl_df/tbl/data.frame)
$ name: chr [1:87] "Luke Skywalker" "C-3PO" "R2-D2" "Darth Vader" ...
$ mass: num [1:87] 77 75 32 136 49 120 75 32 84 77 ...You can also use more general regular expressions. See the help page (?select) for more details.
Subsetting with filter()
The filter() function is used to extract subsets of rows or observations from a data frame. This function is similar to the existing subset() function in base R, or indexing by logical comparisons.

[Source: Artwork by Allison Horst]
Suppose we wanted to extract the rows of the starwars data frame where the birthyear is greater than 100:
# A tibble: 5 × 14
name height mass hair_color skin_color eye_color birth_year sex gender
<chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 C-3PO 167 75 <NA> gold yellow 112 none mascu…
2 Chewbacca 228 112 brown unknown blue 200 male mascu…
3 Jabba De… 175 1358 <NA> green-tan… orange 600 herm… mascu…
4 Yoda 66 17 white green brown 896 male mascu…
5 Dooku 193 80 white fair brown 102 male mascu…
# ℹ 5 more variables: homeworld <chr>, species <chr>, films <list>,
# vehicles <list>, starships <list>You can see that there are now only 5 rows in the data frame and the distribution of the birth_year values is.
summary(age100$birth_year) Min. 1st Qu. Median Mean 3rd Qu. Max.
102 112 200 382 600 896 We can also filter on multiple conditions: and requires both conditions to be true, whereas or requires only one to be true. This time letʻs choose birth_year < 100 and homeworld == "Tatooine:
age_tat <- filter(starwars, birth_year < 100 & homeworld == "Tatooine")
select(age_tat, name, height, mass, birth_year, sex)# A tibble: 8 × 5
name height mass birth_year sex
<chr> <int> <dbl> <dbl> <chr>
1 Luke Skywalker 172 77 19 male
2 Darth Vader 202 136 41.9 male
3 Owen Lars 178 120 52 male
4 Beru Whitesun lars 165 75 47 female
5 Biggs Darklighter 183 84 24 male
6 Anakin Skywalker 188 84 41.9 male
7 Shmi Skywalker 163 NA 72 female
8 Cliegg Lars 183 NA 82 male Other logical operators you should be aware of include:
| Operator | Meaning | Example |
|---|---|---|
== |
Equals | homeworld == Tatooine |
!= |
Does not equal | homeworld != Tatooine |
> |
Greater than | height > 170.0 |
>= |
Greater than or equal to | height >= 170.0 |
< |
Less than | height < 170.0 |
<= |
Less than or equal to | height <= 170.0 |
%in% |
Included in | homeworld %in% c("Tatooine", "Naboo") |
is.na() |
Is a missing value | is.na(mass) |
If you are ever unsure of how to write a logical statement, but know how to write its opposite, you can use the ! operator to negate the whole statement.
A common use of this is to identify observations with non-missing data (e.g., !(is.na(homweworld))).
Sorting data with arrange()
arrange() is like the sort function in a spreadsheet, or order() in base R. arrange() reorders rows of a data frame according to one of the columns. Think of this as sorting your rows on the value of a column.
Here we can order the rows of the data frame by birth_year, so that the first row is the earliest (oldest) observation and the last row is the latest (most recent) observation.
starwars <- arrange(starwars, birth_year)We can now check the first few rows
# A tibble: 3 × 2
name birth_year
<chr> <dbl>
1 Wicket Systri Warrick 8
2 IG-88 15
3 Luke Skywalker 19and the last few rows.
# A tibble: 3 × 2
name birth_year
<chr> <dbl>
1 Poe Dameron NA
2 BB8 NA
3 Captain Phasma NAColumns can be arranged in descending order using the helper function desc().
Looking at the first three and last three rows shows the dates in descending order.
Rearranging columns with relocate()
Moving a column to a new location is done by specifying the column names, and indicating where they go with the .before= or .after= arguments specifing a location (another column).
relocate(.data, ..., .before = NULL, .after = NULL)
Renaming columns with rename()
Renaming a variable in a data frame in R is accomplished using the names() function. The rename() function is designed to make this process easier.
Here you can see the names of the first six variables in the starwars data frame.
head(starwars[, 1:6], 3)# A tibble: 3 × 6
name height mass hair_color skin_color eye_color
<chr> <int> <dbl> <chr> <chr> <chr>
1 Yoda 66 17 white green brown
2 Jabba Desilijic Tiure 175 1358 <NA> green-tan, brown orange
3 Chewbacca 228 112 brown unknown blue Suppose we wanted to drop the _color. The syntax is newname = oldname:
starwars <- rename(starwars, hair = hair_color, skin = skin_color, eye = eye_color)
head(starwars[, 1:6], 3)# A tibble: 3 × 6
name height mass hair skin eye
<chr> <int> <dbl> <chr> <chr> <chr>
1 Yoda 66 17 white green brown
2 Jabba Desilijic Tiure 175 1358 <NA> green-tan, brown orange
3 Chewbacca 228 112 brown unknown blue How would you do the equivalent in base R without dplyr?
Adding columns with mutate()
The mutate() function computes transformations of variables in a data frame.

[Source: Artwork by Allison Horst]
For example, we may want to adjust height for mass:
# A tibble: 6 × 15
name height mass hair skin eye birth_year sex gender homeworld species
<chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr>
1 Yoda 66 17 white green brown 896 male mascu… <NA> Yoda's…
2 Jabb… 175 1358 <NA> gree… oran… 600 herm… mascu… Nal Hutta Hutt
3 Chew… 228 112 brown unkn… blue 200 male mascu… Kashyyyk Wookiee
4 C-3PO 167 75 <NA> gold yell… 112 none mascu… Tatooine Droid
5 Dooku 193 80 white fair brown 102 male mascu… Serenno Human
6 Qui-… 193 89 brown fair blue 92 male mascu… <NA> Human
# ℹ 4 more variables: films <list>, vehicles <list>, starships <list>,
# heightsize <dbl>There is also the related transmute() function, which mutate()s and keeps only the transformed variables. Therefore, the result is only two columns in the transmuted data frame.
Perform functions on groups using group_by()
The group_by() function is used to indicate groups within the data.
For example, what is the average height by homeworld?
In conjunction with the group_by() function, we often use the summarize() function.
The general operation here is a combination of
- Splitting a data frame by group defined by a variable or group of variables (
group_by()) -
summarize()across those subsets
We can create a separate data frame that splits the original data frame by homeworld.
worlds <- group_by(starwars, homeworld)Compute summary statistics by planet (just showing mean and median here, almost any summary stat is available):
summarize(worlds, height = mean(height, na.rm = TRUE),
maxheight = max(height, na.rm = TRUE),
mass = median(mass, na.rm = TRUE))# A tibble: 49 × 4
homeworld height maxheight mass
<chr> <dbl> <dbl> <dbl>
1 Alderaan 176. 176. 64
2 Aleen Minor 79 79 15
3 Bespin 175 175 79
4 Bestine IV 180 180 110
5 Cato Neimoidia 191 191 90
6 Cerea 198 198 82
7 Champala 196 196 NA
8 Chandrila 150 150 NA
9 Concord Dawn 183 183 79
10 Corellia 175 175 78.5
# ℹ 39 more rowssummarize() returns a data frame with homeworld as the first column, followed by the requested summary statistics. This is similar to the base R function aggregate().
In a slightly more complicated example, we might want to know what are the average masses within quintiles of height:
First, we can create a categorical variable of height5 divided into quintiles
Now we can group the data frame by the height.quint variable.
quint <- group_by(starwars, height.quint)Finally, we can compute the mean of mass within quintiles of height.
Oddly enough there is a maximum mass in the second height quintile of Starwars characters. The biologist in me thinks maybe outliers?
Piping multiple functions using %>%
The pipe operator %>% is very handy for stringing together multiple dplyr functions in a sequence of operations. It comes from the magritter package.
In base R, there are two styles of applying multiple functions. The first is the resave the object after each operation.
The second is to nest functions, with the first at the deepest level (the heart of the onion), then working our way out:
third(second(first(x)))The %>% operator allows you to string operations in a left-to-right fashion, where the output of one flows into the next, i.e.:
Take the example that we just did in the last section.
That can be done with the following sequence:
starwars %>%
group_by(homeworld) %>%
summarize(height = mean(height, na.rm = TRUE),
maxheight = max(height, na.rm = TRUE),
mass = median(mass, na.rm = TRUE))# A tibble: 49 × 4
homeworld height maxheight mass
<chr> <dbl> <dbl> <dbl>
1 Alderaan 176. 176. 64
2 Aleen Minor 79 79 15
3 Bespin 175 175 79
4 Bestine IV 180 180 110
5 Cato Neimoidia 191 191 90
6 Cerea 198 198 82
7 Champala 196 196 NA
8 Chandrila 150 150 NA
9 Concord Dawn 183 183 79
10 Corellia 175 175 78.5
# ℹ 39 more rowsData masking
Notice that we did not have to specify the dataframe. This is because dplyr functions are built on a data masking syntax. From the dplyr data-masking help page:
Data masking allows you to refer to variables in the “current” data frame (usually supplied in the .data argument), without any other prefix. It’s what allows you to type (e.g.)
filter(diamonds, x == 0 & y == 0 & z == 0)instead ofdiamonds[diamonds$x == 0 & diamonds$y == 0 & diamonds$z == 0, ]
When you look at the help page for ?mutate for example, you will see a function definition like so:
mutate(.data, ...)
Note the .data, Which means that the data can be supplied as usual, or it can be inherited from the “current” data frame which is passed to it via a pipe.
Sample rows of data with slice_*()
The slice_sample() function will randomly sample rows of data.
The number of rows to show is specified by the n argument.
- This can be useful if you do not want to print the entire tibble, but you want to get a greater sense of the variation.
slice_sample(starwars, n = 10)# A tibble: 10 × 16
name height mass hair skin eye birth_year sex gender homeworld
<chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
1 Darth Vader 202 136 none white yell… 41.9 male mascu… Tatooine
2 San Hill 191 NA none grey gold NA male mascu… Muunilin…
3 Bail Presto… 191 NA black tan brown 67 male mascu… Alderaan
4 Mon Mothma 150 NA aubu… fair blue 48 fema… femin… Chandrila
5 Zam Wesell 168 55 blon… fair… yell… NA fema… femin… Zolan
6 Anakin Skyw… 188 84 blond fair blue 41.9 male mascu… Tatooine
7 Cliegg Lars 183 NA brown fair blue 82 male mascu… Tatooine
8 Sebulba 112 40 none grey… oran… NA male mascu… Malastare
9 Quarsh Pana… 183 NA black dark brown 62 <NA> <NA> Naboo
10 Gregar Typho 185 85 black dark brown NA male mascu… Naboo
# ℹ 6 more variables: species <chr>, films <list>, vehicles <list>,
# starships <list>, heightsize <dbl>, height.quint <fct>You can also use slice_head() or slice_tail() to take a look at the top rows or bottom rows of your tibble. Again the number of rows can be specified with the n argument.
This will show the first 5 rows.
slice_head(starwars, n = 5)# A tibble: 5 × 16
name height mass hair skin eye birth_year sex gender homeworld species
<chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr>
1 Yoda 66 17 white green brown 896 male mascu… <NA> Yoda's…
2 Jabb… 175 1358 <NA> gree… oran… 600 herm… mascu… Nal Hutta Hutt
3 Chew… 228 112 brown unkn… blue 200 male mascu… Kashyyyk Wookiee
4 C-3PO 167 75 <NA> gold yell… 112 none mascu… Tatooine Droid
5 Dooku 193 80 white fair brown 102 male mascu… Serenno Human
# ℹ 5 more variables: films <list>, vehicles <list>, starships <list>,
# heightsize <dbl>, height.quint <fct>This will show the last 5 rows.
slice_tail(starwars, n = 5)# A tibble: 5 × 16
name height mass hair skin eye birth_year sex gender homeworld species
<chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr>
1 Finn NA NA black dark dark NA male mascu… <NA> Human
2 Rey NA NA brown light hazel NA fema… femin… <NA> Human
3 Poe … NA NA brown light brown NA male mascu… <NA> Human
4 BB8 NA NA none none black NA none mascu… <NA> Droid
5 Capt… NA NA unkn… unkn… unkn… NA <NA> <NA> <NA> <NA>
# ℹ 5 more variables: films <list>, vehicles <list>, starships <list>,
# heightsize <dbl>, height.quint <fct>Summary
The dplyr package provides an alternative syntax for manipulating data frames. In particular, we can often conduct the beginnings of an exploratory analysis with the powerful combination of group_by() and summarize().
Once you learn the dplyr grammar there are a few additional benefits
dplyrcan work with other data frame “back ends” such as SQL databases. There is an SQL interface for relational databases via the DBI packagedplyrcan be integrated with thedata.tablepackage for large fast tablesMany people like the piping syntax for readability and clarity
Post-lecture materials
Final Questions
- How can you tell if an object is a tibble?
- Using the
treesdataset in base R (this dataset stores the girth, height, and volume for Black Cherry Trees) and using the pipe operator:- convert the
data.frameto a tibble. - filter for rows with a tree height of greater than 70, and
- order rows by
Volume(smallest to largest).
- convert the
head(trees) Girth Height Volume
1 8.3 70 10.3
2 8.6 65 10.3
3 8.8 63 10.2
4 10.5 72 16.4
5 10.7 81 18.8
6 10.8 83 19.7